Flight tracks#
The P3 flew 95 hours of observations over eleven flights, many of which were coordinated with the NOAA research ship R/V Ronald H. Brown and autonomous platforms deployed from the ship. Each flight contained a mixture of sampling strategies including: high-altitude circles with frequent dropsonde deployment to characterize the large-scale environment; slow descents and ascents to measure the distribution of water vapor and its isotopic composition; stacked legs aimed at sampling the microphysical and thermodynamic state of the boundary layer; and offset straight flight legs for observing clouds and the ocean surface with remote sensing instruments and the thermal structure of the ocean with in situ sensors dropped from the plane.
As a result of this diverse sampling the flight tracks are much more variable then for most of the other aircraft.
General setup:
import xarray as xr
import numpy as np
import datetime
#
# Related to plotting
#
import matplotlib.pyplot as plt
import pathlib
plt.style.use(pathlib.Path("./mplstyle/book"))
%matplotlib inline
Now access the flight track data.
import eurec4a
cat = eurec4a.get_intake_catalog(use_ipfs="QmahMN2wgPauHYkkiTGoG2TpPBmj3p5FoYJAq9uE9iXT9N")
Mapping takes quite some setup. Maybe we’ll encapsulate this later but for now we repeat code in each notebook.
What days did the P-3 fly on? We can find out via the flight segmentation files.
# On what days did the P-3 fly? These are UTC date
all_flight_segments = eurec4a.get_flight_segments()
flight_dates = np.unique([np.datetime64(flight["takeoff"]).astype("datetime64[D]")
for flight in all_flight_segments["P3"].values()])
Now set up colors to code each flight date during the experiment. One could choose a categorical palette so the colors were as different from each other as possible. Here we’ll choose from a continuous set that spans the experiment so days that are close in time are also close in color.
For plotting purposes it’ll be handy to define a one-day time window and to convert between date/time formats
one_day = np.timedelta64(1, "D")
def to_datetime(dt64):
epoch = np.datetime64("1970-01-01")
second = np.timedelta64(1, "s")
timestamp = (dt64 - epoch) / second
return datetime.datetime.fromtimestamp(timestamp, datetime.UTC)
Most platforms available from the EUREC⁴A intake catalog have a tracks element but
we’ll use the flight-level data instead. We’re using xr.concat() with one dask array per day
to avoid loading the whole dataset into memory at once.
nav_data = xr.concat([entry.to_dask().chunk() for entry in cat.P3.flight_level.values()],
dim = "time")
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
A map showing each of the eleven flight tracks:
fig = plt.figure(figsize = (12,13.6))
ax = set_up_map(plt)
add_gridlines(ax)
for d in flight_dates:
flight = nav_data.sel(time=slice(d, d + one_day))
ax.plot(flight.lon, flight.lat,
lw=2, alpha=0.5, c=color_of_day(d),
transform=ccrs.PlateCarree(), zorder=7,
label=f"{to_datetime(d):%m-%d}")
plt.legend(ncol=3, loc=(0.0,0.0), fontsize=14, framealpha=0.8, markerscale=5,
title="Flight date (MM-DD-2020)")
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
/home/runner/miniconda3/envs/how_to_eurec4a/lib/python3.13/site-packages/shapely/creation.py:218: RuntimeWarning: invalid value encountered in linestrings
return lib.linestrings(coords, np.intc(handle_nan), out=out, **kwargs)
<matplotlib.legend.Legend at 0x7f493c4dc2f0>
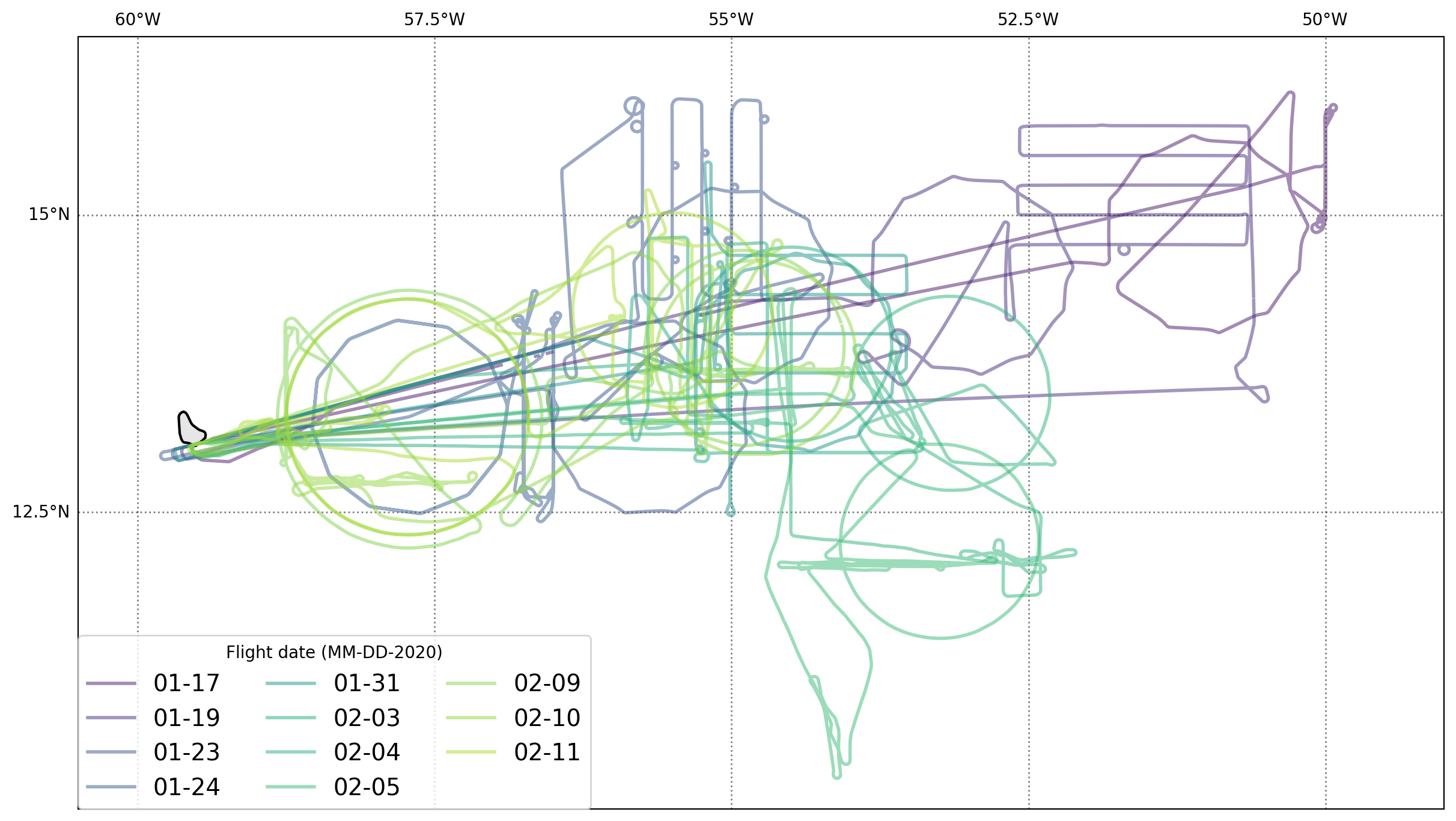
Most dropsondes were deployed from regular dodecagons during the first part of the experiment with short turns after each dropsonde providing an off-nadir look at the ocean surface useful for calibrating the W-band radar. A change in pilots midway through the experiment led to dropsondes being deployed from circular flight tracks starting on 31 Jan. AXBTs were deployed in lawnmower patterns (parallel offset legs) with small loops sometimes employed to lengthen the time between AXBT deployment to allow time for data acquisition given the device’s slow fall speeds. Profiling and especially in situ cloud sampling legs sometimes deviated from straight paths to avoid hazardous weather.
Side view using the same color scale:
fig = plt.figure(figsize = (8.3,5))
ax = plt.axes()
for d in flight_dates:
flight = nav_data.sel(time=slice(d, d + one_day)).compute()
flight = flight.where(flight.alt > 80, drop=True) # Proxy for take-off time
plt.plot(flight.time - flight.time.min(), flight.alt/1000.,
lw=2, alpha=0.5, c=color_of_day(d),
label=f"{to_datetime(d):%m-%d}")
plt.xticks(np.arange(10) * 3600 * 1e9, labels = np.arange(10))
ax.set_xlabel("Time after flight start (h)")
ax.set_ylabel("Aircraft altitude (km)")
Text(0, 0.5, 'Aircraft altitude (km)')
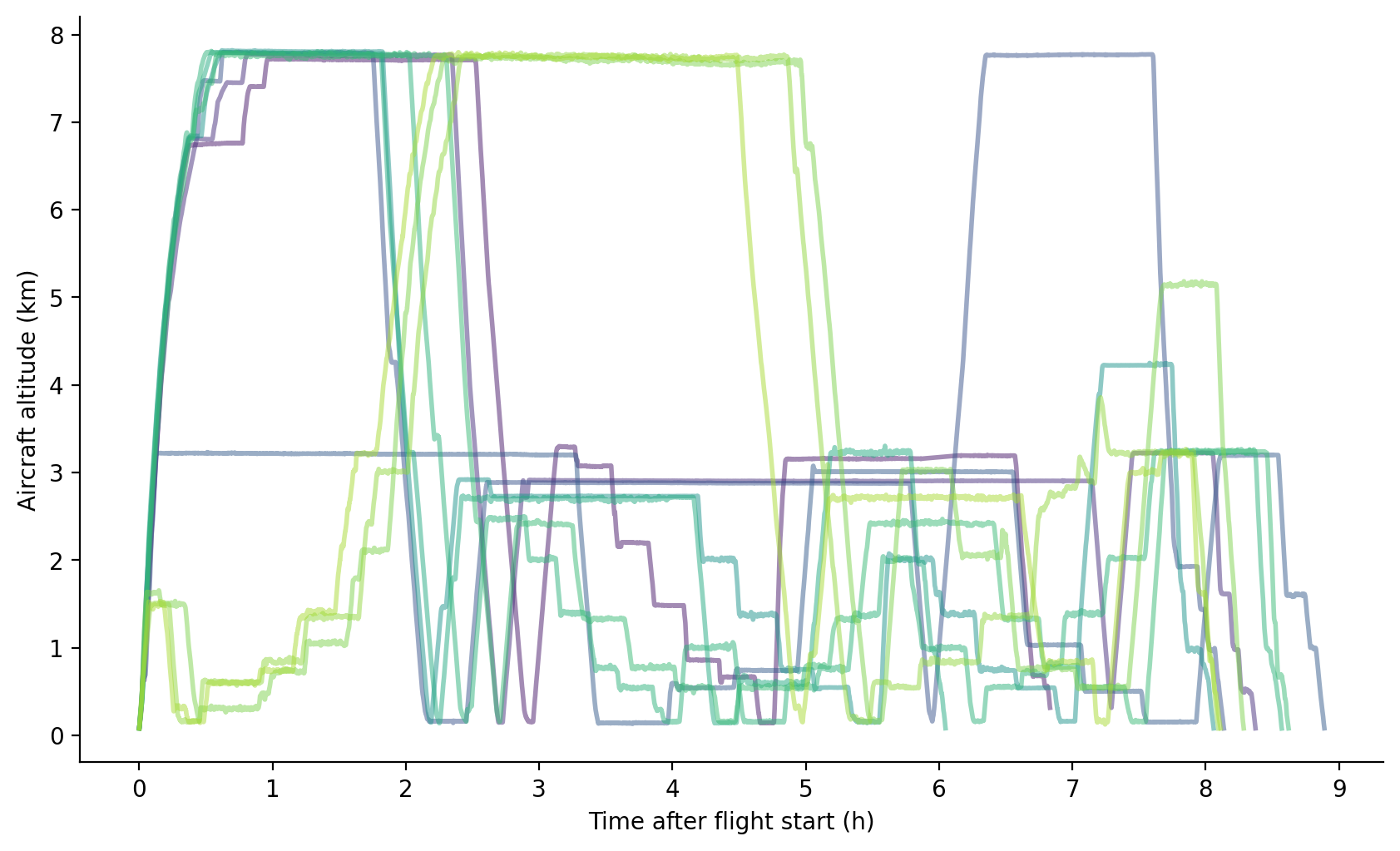
Sondes were dropped from the P-3 at about 7.5 km, with each circle taking roughly an hour; transits were frequently performed at this level to conserve fuel. Long intervals near 3 km altitude were used to deploy AXBTs and/or characterize the ocean surface with remote sensing. Stepped legs indicate times devoted to in situ cloud sampling. On most flights the aircraft climbed quickly to roughly 7.5 km, partly to deconflict with other aircraft participating in the experiment. On the three night flights, however, no other aircraft were operating at take-off times and cloud sampling was performed first, nearer Barbados than on other flights.


