The issue with netCDF data types#
TL; DR
If you want to be on the safe side, use only SHORT, INT, FLOAT, DOUBLE, STRING or array of CHAR as data types.
In particular, don’t use single byte integers, any unsigned integer and no 64 bit integer.
The data format netCDF which is used in many places throughout our community has evolved over time. During this evolution, the basic data types representable within the netCDF data format have been expanded. Currently we are at netCDF version 4 with HDF5 as a storage backend, which offers a lot more flexibility compared to previous versions of netCDF including many more data types. One could assume that this is all fine and we can use these data types freely, but here we are unlucky and there is more to keep in mind.
In order to paint the full picture, we’ll have to take a little detour and think about what netCDF actually is or even more what we think (or probably should think) about when talking about netCDF. As we’ll see, this is not something which can be answered in a straight forward manner, so it is fair to ask if we should care at all. Finally we’ll have a look at an experiment which shows plenty of ways in which different data types may fail around netCDF and which types seem to be fine.
What is netCDF?#
NetCDF is more than only the “netCDF4 on HDF5” storage format. This is not only a bold statement, but is actually asked and discussed by the netCDF developers (What is netCDF?, Jan 2020). This chapter is not exactly about the same topic in the referenced issue, but still has similar roots. The relevant part for this discussion is that there is at least netCDF, the data model and netCDF, the persistence format. It is also recognized that there are multiple persistence formats (netCDF3, netCDF4/HDF5, zarr) for the same data model and there are translation layers such as OPeNDAP which can be used as a transport mechanism between the computer on which data is stored and the computer on which data is analyzed. We further look at the relevance of the netCDF data model and netCDF persistence format for the EUREC⁴A datasets.
The data model(s)#
There are actually a few different ideas of data models around, which share some general ideas. Maybe the most illustrative way to show this general idea is an extended version of xarray’s logo, but the underlying ideas are of course applicable independently of xarray or even Python.
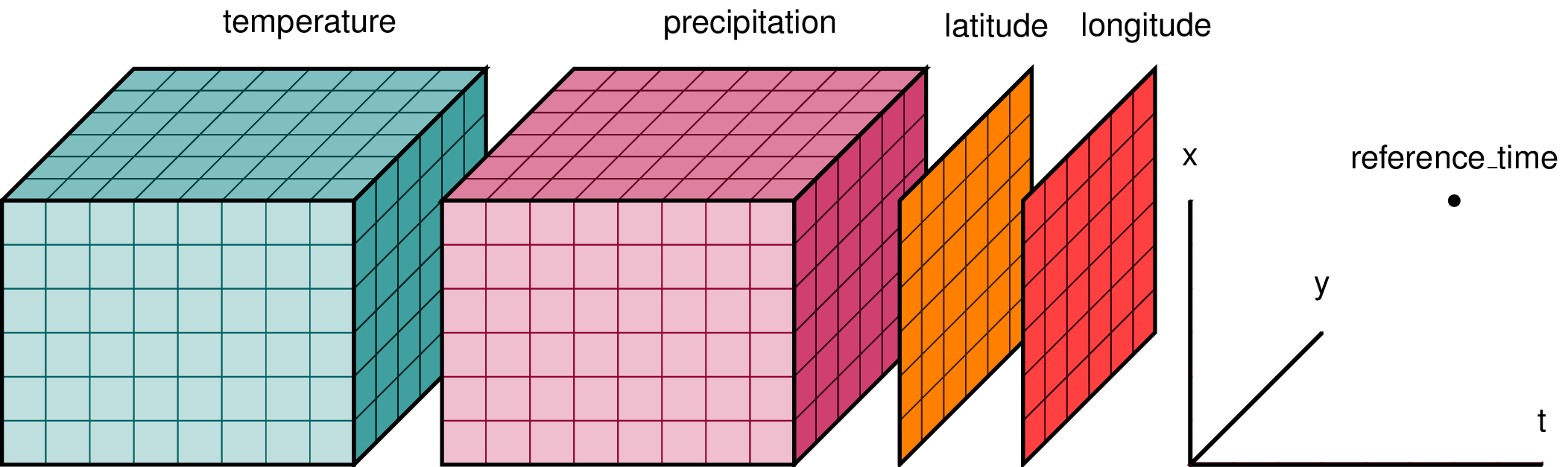
Fig. 4 Dataset diagram (From the xarray documentation)#
A dataset is a collection of variables and dimensions.
Variables are multi-dimensional arrays and they can share dimensions (i.e. to express that temperature and precipitation are defined at the same locations, the three axes of the corresponding arrays should be identified with the same dimension label, that is e.g. x, y and z).
Some variables may be promoted into a coordinate, which does not require to store it differently, but it changes the interpretation of the data.
In the example above, latitude and longitude would likely be identified as coordinates.
One would typically use coordinate values to index into some data or to label the ticks on a plot axis, while a normal variable would usually be used to define a line or a color within a plot.
There may be variables which should be coordinates only for a specific use case and vice versa.
It is useful to distinguish between variables and coordinates within the metadata of a dataset, but the representation of the actual data may be the same for both, variables and coordinates.
In addition to the existence of coordinates, there are more pieces of valuable additional information (think of units, coordinate reference systems, author information etc…).
Thus we usually want those extra bits to datasets and variables, which can be done by the use of attributes.
It turns out that many datasets in the EUREC⁴A context can be represented very well in this model. I assume that in many cases when we talk about that a dataset being available in netCDF, what we really care about is that the dataset is organized along this general structure and thus can be accesses as if it were netCDF.
The storage and transport formats#
netCDF#
In order to put the high level idea from above into a data structure which can be stored on disk, netCDF defines two low level data models where the Classic data model came first and the Enhanced data model evolved from it. Both models care about variables, dimensions and attributes as introduced above.
The classic model knows about CHAR, BYTE, SHORT, INT, FLOAT and DOUBLE data types which are all signed.
There may be at most one unlimited (i.e. resizeable) dimension per variable.
The enhanced model is based on the classic model and adds INT64 and STRING to the mix as well as unsigned variants of all integral types.
Further additions of the enhanced model are groups (which are like a dataset within a dataset) and user defined types.
There may be any number of unlimited dimensions per variable.
Those two data models are closely tied to the netCDF3 and netCDF4 storage formats.
netCDF3 is based on the classic data model and defines a storage format which is custom to netCDF.
netCDF4 is based on the enhanced data model and uses HDF5 as its backend storage format. HDF5 enables further enhancements like internal data compression.
Both storage formats store all data in a single file, chunking of the dataset is done internally. If the dataset should be accessed from a remote location, usually the entire dataset must be fetched before accessing the data.
In Jan 2019 HTTP byte-range enabled partial reads from remote datasets have been added to netCDF-c, but as stated in the pull request, it is not meant for production use:
Note that this is not intended as a true production capability because, as is known, this kind of access to can be quite slow. In addition, the byte-range IO drivers do not currently do any sort of optimization or caching.
Thus, if a dataset should be accessed efficiently from a remote location, something different must be added, which is what OPeNDAP is for.
OPeNDAP#
OPeNDAP is a network protocol to access scientific data via network. In particular, it uses HTTP as a transport mechanism and defines a way to request subsets of a dataset which is formed along the data model as described above. OPeNDAP is thus the go-to method if an existing netCDF dataset should be made available remotely. In the context of EUREC⁴A, most datacenters provide access to uploaded datasets via OPeNDAP.
OPeNDAP is specified in two versions, namely version 2 and version 4. However, DAP4 is still a draft since 2004 and the only widely supported version of OPeNDAP is version 2.
The data model of OPeNDAP v2 is slightly different from the data model in netCDF.
Regarding data types, OPeNDAP v2 defines Byte, Int16, UInt16, Int32, UInt32, Float32, Float64, String, URL.
As opposed to the BYTE in netCDF Classic, the Byte in OPeNDAP is unsigned.
This makes byte types of netCDF Classic and OPeNDAP incompatible, but as noted in the summary, there exists a hack which tries to circumvent that.
One could assume that UBYTE of netCDF Enhanced would fit to this Byte, but there are issues as well which are confirmed in the experiment.
Furthermore, there is no CHAR, so whenever a netCDF dataset containing text as a sequence of CHAR is encountered by an OPeNDAP server, this will be converted to an OPeNDAP String.
A consequence of this is that CHARs and STRINGs can not be distinguished when transferred over OPeNDAP and thus may be converted into each other after one cycle through netCDF → OPeNDAP → netCDF.
This behaviour is not (yet) studied in this text.
zarr#
to do
zarr should be discussed in this place.
Why should we care?#
Each of the formats has distinct advantages and disadvantages:
netCDF is a compact format, the whole dataset can be stored in a single file which can be transferred from one place to another and everything is contained. This is great for storing data and to send around smaller datasets, but retrieving only a subset of a dataset from a remote server is not possible. Libraries which can open netCDF files usually do not handle HTTP, so the dataset needs to be stored in a local directory to use it.
OPeNDAP is a network protocol based on HTTP which has been made exactly for the purpose of requesting subsets of a dataset from a remote server. As it is only a network protocol, it can not be used to store the data, so every dataset which should be provided via OPeNDAP must be stored in another format (like netCDF) or computed on the fly. Support for reading OPeNDAP is build into netCDF-c.
zarr is a storage format which is spread out into several files in a defined directory structure. This makes it a little harder to pass around, but this chunked structure makes remote access exceptionally simple and fast. Chelle Gentemann has prepared an impressive demonstration on this topic.
So, depending on the use case, different formats are optimal and none of them supports everything:
format |
for storage |
sending around |
for remote access |
widely supported |
|---|---|---|---|---|
netCDF |
✅ |
✅ |
❌, works via OPeNDAP |
✅ |
OPeNDAP |
❌ |
❌ |
✅, can share netCDF |
✅ |
zarr |
✅ |
moderate |
✅, with high performance |
❌, netCDF-c is working on it |
Thus, if we want to have datasets which can be used locally as well as remotely (some might call it “in the cloud”), we should take care that our datasets are convertible between those formats so that we can adapt to the specific use case.
An experiment#
Currently, the main use case for the Aeris data server is to publish netCDF files and retrieve them back, either via direct download or via OPeNDAP. This experiment is designed to test under which circumstances data which has been created as netCDF can be retrieved correctly via OPeNDAP. In order to minimize additional problems due to the mix of incompatible libraries, all dataset decoding is done using the same netCDF-c library (independent of the access to the dataset, directly or via OPeNDAP).
The individual test cases show differences between data types, the use of negative numbers, the use of values used as numeric values, and the use of values interpreted as flags. For each data type, there is a drop down menu which shows the individual sub cases. If any one of the sub cases failed, the data type is marked as erroneous.
In each case the test follows the same steps:
The original dataset is dumped via
ncdumpThe OPeNDAP dataset attribute structure (
.das) is retrieved viacurlto look at the raw response of the serverThe dataset as received via OPeNDAP is dumped via
ncdumpA little script checks if it detects any significant error between the original and the OPeNDAP dataset
Results#
BYTE
original dataset:
$ ncdump test_NC_BYTE.nc
netcdf test_NC_BYTE {
dimensions:
test = 5 ;
variables:
byte test(test) ;
test:_FillValue = 3b ;
test:valid_range = 0b, 127b ;
data:
test = 0, 1, 2, _, 127 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE.nc.das
Attributes {
test {
String _Unsigned "false";
Byte _FillValue 3;
Byte valid_range 0, 127;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE.nc
netcdf test_NC_BYTE {
dimensions:
test = 5 ;
variables:
byte test(test) ;
test:_Unsigned = "false" ;
test:_FillValue = 3b ;
test:valid_range = 0b, 127b ;
data:
test = 0, 1, 2, _, 127 ;
}
✅
original dataset:
$ ncdump test_NC_BYTE_neg.nc
netcdf test_NC_BYTE_neg {
dimensions:
test = 7 ;
variables:
byte test(test) ;
test:_FillValue = 3b ;
test:valid_range = -128b, 127b ;
data:
test = -128, -1, 0, 1, 2, _, 127 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_neg.nc.das
Attributes {
test {
String _Unsigned "false";
Byte _FillValue 3;
Int16 valid_range -128, 127;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_neg.nc
netcdf test_NC_BYTE_neg {
dimensions:
test = 7 ;
variables:
byte test(test) ;
test:_Unsigned = "false" ;
test:_FillValue = 3b ;
test:valid_range = -128s, 127s ;
data:
test = -128, -1, 0, 1, 2, _, 127 ;
}
Error
data type of attribute valid_range of variable test differs
original dataset:
$ ncdump test_NC_BYTE_flag.nc
netcdf test_NC_BYTE_flag {
dimensions:
test = 5 ;
variables:
byte test(test) ;
test:flag_values = 0b, 1b, 2b, 3b, 127b ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 127 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_flag.nc.das
Attributes {
test {
String _Unsigned "false";
Byte flag_values 0, 1, 2, 3, 127;
String flag_meanings "a b c d e";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_flag.nc
netcdf test_NC_BYTE_flag {
dimensions:
test = 5 ;
variables:
byte test(test) ;
test:_Unsigned = "false" ;
test:flag_values = 0b, 1b, 2b, 3b, 127b ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 127 ;
}
✅
original dataset:
$ ncdump test_NC_BYTE_flag_neg.nc
netcdf test_NC_BYTE_flag_neg {
dimensions:
test = 7 ;
variables:
byte test(test) ;
test:flag_values = -128b, -1b, 0b, 1b, 2b, 3b, 127b ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -128, -1, 0, 1, 2, 3, 127 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_flag_neg.nc.das
Attributes {
test {
String _Unsigned "false";
Int16 flag_values -128, -1, 0, 1, 2, 3, 127;
String flag_meanings "a b c d e f g";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_BYTE_flag_neg.nc
netcdf test_NC_BYTE_flag_neg {
dimensions:
test = 7 ;
variables:
byte test(test) ;
test:_Unsigned = "false" ;
test:flag_values = -128s, -1s, 0s, 1s, 2s, 3s, 127s ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -128, -1, 0, 1, 2, 3, 127 ;
}
Error
data type of attribute flag_values of variable test differs
UBYTE
original dataset:
$ ncdump test_NC_UBYTE.nc
netcdf test_NC_UBYTE {
dimensions:
test = 5 ;
variables:
ubyte test(test) ;
test:_FillValue = 3UB ;
test:valid_range = 0UB, 255UB ;
data:
test = 0, 1, 2, _, 255 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE.nc.das
Attributes {
test {
String _Unsigned "true";
Byte _FillValue 3;
Int16 valid_range 0, -1;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE.nc
netcdf test_NC_UBYTE {
dimensions:
test = 5 ;
variables:
byte test(test) ;
test:_Unsigned = "true" ;
test:_FillValue = 3b ;
test:valid_range = 0s, -1s ;
data:
test = 0, 1, 2, _, -1 ;
}
Error
values of variable test are not equal
original dataset:
$ ncdump test_NC_UBYTE_flag.nc
netcdf test_NC_UBYTE_flag {
dimensions:
test = 5 ;
variables:
ubyte test(test) ;
test:flag_values = 0UB, 1UB, 2UB, 3UB, 255UB ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 255 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE_flag.nc.das
Attributes {
test {
String _Unsigned "true";
Int16 flag_values 0, 1, 2, 3, -1;
String flag_meanings "a b c d e";
Int16 _FillValue -1;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE_flag.nc
Error
/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE_flag.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE_flag.nc: NetCDF: Not a valid data type or _FillValue type mismatch
Error
[Errno -45] NetCDF: Not a valid data type or _FillValue type mismatch: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UBYTE_flag.nc’
SHORT
original dataset:
$ ncdump test_NC_SHORT.nc
netcdf test_NC_SHORT {
dimensions:
test = 5 ;
variables:
short test(test) ;
test:_FillValue = 3s ;
test:valid_range = 0s, 32767s ;
data:
test = 0, 1, 2, _, 32767 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT.nc.das
Attributes {
test {
Int16 _FillValue 3;
Int16 valid_range 0, 32767;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT.nc
netcdf test_NC_SHORT {
dimensions:
test = 5 ;
variables:
short test(test) ;
test:_FillValue = 3s ;
test:valid_range = 0s, 32767s ;
data:
test = 0, 1, 2, _, 32767 ;
}
✅
original dataset:
$ ncdump test_NC_SHORT_neg.nc
netcdf test_NC_SHORT_neg {
dimensions:
test = 7 ;
variables:
short test(test) ;
test:_FillValue = 3s ;
test:valid_range = -32768s, 32767s ;
data:
test = -32768, -1, 0, 1, 2, _, 32767 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_neg.nc.das
Attributes {
test {
Int16 _FillValue 3;
Int16 valid_range -32768, 32767;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_neg.nc
netcdf test_NC_SHORT_neg {
dimensions:
test = 7 ;
variables:
short test(test) ;
test:_FillValue = 3s ;
test:valid_range = -32768s, 32767s ;
data:
test = -32768, -1, 0, 1, 2, _, 32767 ;
}
✅
original dataset:
$ ncdump test_NC_SHORT_flag.nc
netcdf test_NC_SHORT_flag {
dimensions:
test = 5 ;
variables:
short test(test) ;
test:flag_values = 0s, 1s, 2s, 3s, 32767s ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 32767 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_flag.nc.das
Attributes {
test {
Int16 flag_values 0, 1, 2, 3, 32767;
String flag_meanings "a b c d e";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_flag.nc
netcdf test_NC_SHORT_flag {
dimensions:
test = 5 ;
variables:
short test(test) ;
test:flag_values = 0s, 1s, 2s, 3s, 32767s ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 32767 ;
}
✅
original dataset:
$ ncdump test_NC_SHORT_flag_neg.nc
netcdf test_NC_SHORT_flag_neg {
dimensions:
test = 7 ;
variables:
short test(test) ;
test:flag_values = -32768s, -1s, 0s, 1s, 2s, 3s, 32767s ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -32768, -1, 0, 1, 2, 3, 32767 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_flag_neg.nc.das
Attributes {
test {
Int16 flag_values -32768, -1, 0, 1, 2, 3, 32767;
String flag_meanings "a b c d e f g";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_SHORT_flag_neg.nc
netcdf test_NC_SHORT_flag_neg {
dimensions:
test = 7 ;
variables:
short test(test) ;
test:flag_values = -32768s, -1s, 0s, 1s, 2s, 3s, 32767s ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -32768, -1, 0, 1, 2, 3, 32767 ;
}
✅
USHORT
original dataset:
$ ncdump test_NC_USHORT.nc
netcdf test_NC_USHORT {
dimensions:
test = 5 ;
variables:
ushort test(test) ;
test:_FillValue = 3US ;
test:valid_range = 0US, 65535US ;
data:
test = 0, 1, 2, _, 65535 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT.nc.das
Attributes {
test {
Int16 _FillValue 3;
Int16 valid_range 0, -1;
String _Unsigned "true";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT.nc
Error
/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT.nc: NetCDF: Not a valid data type or _FillValue type mismatch
Error
[Errno -45] NetCDF: Not a valid data type or _FillValue type mismatch: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT.nc’
original dataset:
$ ncdump test_NC_USHORT_flag.nc
netcdf test_NC_USHORT_flag {
dimensions:
test = 5 ;
variables:
ushort test(test) ;
test:flag_values = 0US, 1US, 2US, 3US, 65535US ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, _ ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT_flag.nc.das
Attributes {
test {
Int16 flag_values 0, 1, 2, 3, -1;
String flag_meanings "a b c d e";
Int16 _FillValue -1;
String _Unsigned "true";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT_flag.nc
Error
/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT_flag.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT_flag.nc: NetCDF: Not a valid data type or _FillValue type mismatch
Error
[Errno -45] NetCDF: Not a valid data type or _FillValue type mismatch: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_USHORT_flag.nc’
INT
original dataset:
$ ncdump test_NC_INT.nc
netcdf test_NC_INT {
dimensions:
test = 5 ;
variables:
int test(test) ;
test:_FillValue = 3 ;
test:valid_range = 0, 2147483647 ;
data:
test = 0, 1, 2, _, 2147483647 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT.nc.das
Attributes {
test {
Int32 _FillValue 3;
Int32 valid_range 0, 2147483647;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT.nc
netcdf test_NC_INT {
dimensions:
test = 5 ;
variables:
int test(test) ;
test:_FillValue = 3 ;
test:valid_range = 0, 2147483647 ;
data:
test = 0, 1, 2, _, 2147483647 ;
}
✅
original dataset:
$ ncdump test_NC_INT_neg.nc
netcdf test_NC_INT_neg {
dimensions:
test = 7 ;
variables:
int test(test) ;
test:_FillValue = 3 ;
test:valid_range = -2147483648, 2147483647 ;
data:
test = -2147483648, -1, 0, 1, 2, _, 2147483647 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_neg.nc.das
Attributes {
test {
Int32 _FillValue 3;
Int32 valid_range -2147483648, 2147483647;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_neg.nc
netcdf test_NC_INT_neg {
dimensions:
test = 7 ;
variables:
int test(test) ;
test:_FillValue = 3 ;
test:valid_range = -2147483648, 2147483647 ;
data:
test = -2147483648, -1, 0, 1, 2, _, 2147483647 ;
}
✅
original dataset:
$ ncdump test_NC_INT_flag.nc
netcdf test_NC_INT_flag {
dimensions:
test = 5 ;
variables:
int test(test) ;
test:flag_values = 0, 1, 2, 3, 2147483647 ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 2147483647 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_flag.nc.das
Attributes {
test {
Int32 flag_values 0, 1, 2, 3, 2147483647;
String flag_meanings "a b c d e";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_flag.nc
netcdf test_NC_INT_flag {
dimensions:
test = 5 ;
variables:
int test(test) ;
test:flag_values = 0, 1, 2, 3, 2147483647 ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 2147483647 ;
}
✅
original dataset:
$ ncdump test_NC_INT_flag_neg.nc
netcdf test_NC_INT_flag_neg {
dimensions:
test = 7 ;
variables:
int test(test) ;
test:flag_values = -2147483648, -1, 0, 1, 2, 3, 2147483647 ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -2147483648, -1, 0, 1, 2, 3, 2147483647 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_flag_neg.nc.das
Attributes {
test {
Int32 flag_values -2147483648, -1, 0, 1, 2, 3, 2147483647;
String flag_meanings "a b c d e f g";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT_flag_neg.nc
netcdf test_NC_INT_flag_neg {
dimensions:
test = 7 ;
variables:
int test(test) ;
test:flag_values = -2147483648, -1, 0, 1, 2, 3, 2147483647 ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -2147483648, -1, 0, 1, 2, 3, 2147483647 ;
}
✅
UINT
original dataset:
$ ncdump test_NC_UINT.nc
netcdf test_NC_UINT {
dimensions:
test = 5 ;
variables:
uint test(test) ;
test:_FillValue = 3U ;
test:valid_range = 0U, 4294967295U ;
data:
test = 0, 1, 2, _, 4294967295 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT.nc.das
Attributes {
test {
Int32 _FillValue 3;
Int32 valid_range 0, -1;
String _Unsigned "true";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT.nc
Error
/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT.nc: NetCDF: Not a valid data type or _FillValue type mismatch
Error
[Errno -45] NetCDF: Not a valid data type or _FillValue type mismatch: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT.nc’
original dataset:
$ ncdump test_NC_UINT_flag.nc
netcdf test_NC_UINT_flag {
dimensions:
test = 5 ;
variables:
uint test(test) ;
test:flag_values = 0U, 1U, 2U, 3U, 4294967295U ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, _ ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT_flag.nc.das
Attributes {
test {
Int32 flag_values 0, 1, 2, 3, -1;
String flag_meanings "a b c d e";
Int32 _FillValue -1;
String _Unsigned "true";
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT_flag.nc
Error
/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT_flag.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT_flag.nc: NetCDF: Not a valid data type or _FillValue type mismatch
Error
[Errno -45] NetCDF: Not a valid data type or _FillValue type mismatch: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT_flag.nc’
INT64
original dataset:
$ ncdump test_NC_INT64.nc
netcdf test_NC_INT64 {
dimensions:
test = 5 ;
variables:
int64 test(test) ;
test:_FillValue = 3LL ;
test:valid_range = 0LL, 9223372036854775807LL ;
data:
test = 0, 1, 2, _, 9223372036854775807 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64.nc’
original dataset:
$ ncdump test_NC_INT64_neg.nc
netcdf test_NC_INT64_neg {
dimensions:
test = 7 ;
variables:
int64 test(test) ;
test:_FillValue = 3LL ;
test:valid_range = -9223372036854775808LL, 9223372036854775807LL ;
data:
test = -9223372036854775808, -1, 0, 1, 2, _, 9223372036854775807 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_neg.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_neg.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_neg.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_neg.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_neg.nc’
original dataset:
$ ncdump test_NC_INT64_flag.nc
netcdf test_NC_INT64_flag {
dimensions:
test = 5 ;
variables:
int64 test(test) ;
test:flag_values = 0LL, 1LL, 2LL, 3LL, 9223372036854775807LL ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 9223372036854775807 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag.nc’
original dataset:
$ ncdump test_NC_INT64_flag_neg.nc
netcdf test_NC_INT64_flag_neg {
dimensions:
test = 7 ;
variables:
int64 test(test) ;
test:flag_values = -9223372036854775808LL, -1LL, 0LL, 1LL, 2LL, 3LL, 9223372036854775807LL ;
test:flag_meanings = "a b c d e f g" ;
data:
test = -9223372036854775808, -1, 0, 1, 2, 3, 9223372036854775807 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag_neg.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag_neg.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag_neg.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag_neg.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_INT64_flag_neg.nc’
UINT64
original dataset:
$ ncdump test_NC_UINT64.nc
netcdf test_NC_UINT64 {
dimensions:
test = 5 ;
variables:
uint64 test(test) ;
test:_FillValue = 3ULL ;
test:valid_range = 0ULL, 0ULL ;
data:
test = 0, 1, 2, _, 18446744073709551615 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64.nc’
original dataset:
$ ncdump test_NC_UINT64_flag.nc
netcdf test_NC_UINT64_flag {
dimensions:
test = 5 ;
variables:
uint64 test(test) ;
test:flag_values = 0ULL, 1ULL, 2ULL, 3ULL, 0ULL ;
test:flag_meanings = "a b c d e" ;
data:
test = 0, 1, 2, 3, 18446744073709551615 ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64_flag.nc.das
Error {
code = 403;
message = "NcDDS Variable data type = long";
};
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64_flag.nc
Error
oc_open: server error retrieving url: code=403 message=”NcDDS Variable data type = long”/usr/local/bin/ncdump: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64_flag.nc: https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64_flag.nc: NetCDF: Access failure
Error
[Errno -77] NetCDF: Access failure: b’https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_UINT64_flag.nc’
FLOAT
original dataset:
$ ncdump test_NC_FLOAT.nc
netcdf test_NC_FLOAT {
dimensions:
test = 7 ;
variables:
float test(test) ;
test:_FillValue = NaNf ;
test:valid_range = 0.f, Infinityf ;
data:
test = 0, 1, 2, 3, _, 3.402823e+38, Infinityf ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_FLOAT.nc.das
Attributes {
test {
Float32 _FillValue NaN;
Float32 valid_range 0.0, Infinity;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_FLOAT.nc
netcdf test_NC_FLOAT {
dimensions:
test = 7 ;
variables:
float test(test) ;
test:_FillValue = NaNf ;
test:valid_range = 0.f, Infinityf ;
data:
test = 0, 1, 2, 3, _, 3.402823e+38, Infinityf ;
}
✅
original dataset:
$ ncdump test_NC_FLOAT_neg.nc
netcdf test_NC_FLOAT_neg {
dimensions:
test = 10 ;
variables:
float test(test) ;
test:_FillValue = NaNf ;
test:valid_range = -Infinityf, Infinityf ;
data:
test = -Infinityf, -3.402823e+38, -1, 0, 1, 2, 3, _, 3.402823e+38, Infinityf ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_FLOAT_neg.nc.das
Attributes {
test {
Float32 _FillValue NaN;
Float32 valid_range -Infinity, Infinity;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_FLOAT_neg.nc
netcdf test_NC_FLOAT_neg {
dimensions:
test = 10 ;
variables:
float test(test) ;
test:_FillValue = NaNf ;
test:valid_range = -Infinityf, Infinityf ;
data:
test = -Infinityf, -3.402823e+38, -1, 0, 1, 2, 3, _, 3.402823e+38, Infinityf ;
}
✅
DOUBLE
original dataset:
$ ncdump test_NC_DOUBLE.nc
netcdf test_NC_DOUBLE {
dimensions:
test = 7 ;
variables:
double test(test) ;
test:_FillValue = NaN ;
test:valid_range = 0., Infinity ;
data:
test = 0, 1, 2, 3, _, 1.79769313486232e+308, Infinity ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_DOUBLE.nc.das
Attributes {
test {
Float64 _FillValue NaN;
Float64 valid_range 0.0, Infinity;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_DOUBLE.nc
netcdf test_NC_DOUBLE {
dimensions:
test = 7 ;
variables:
double test(test) ;
test:_FillValue = NaN ;
test:valid_range = 0., Infinity ;
data:
test = 0, 1, 2, 3, _, 1.79769313486232e+308, Infinity ;
}
✅
original dataset:
$ ncdump test_NC_DOUBLE_neg.nc
netcdf test_NC_DOUBLE_neg {
dimensions:
test = 10 ;
variables:
double test(test) ;
test:_FillValue = NaN ;
test:valid_range = -Infinity, Infinity ;
data:
test = -Infinity, -1.79769313486232e+308, -1, 0, 1, 2, 3, _,
1.79769313486232e+308, Infinity ;
}
data attribute structure via OPeNDAP:
$ curl https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_DOUBLE_neg.nc.das
Attributes {
test {
Float64 _FillValue NaN;
Float64 valid_range -Infinity, Infinity;
}
}
dataset as interpreted by netCDF via OPeNDAP:
$ ncdump https://observations.ipsl.fr/thredds/dodsC/EUREC4A/PRODUCTS/testdata/netcdf_testfiles/test_NC_DOUBLE_neg.nc
netcdf test_NC_DOUBLE_neg {
dimensions:
test = 10 ;
variables:
double test(test) ;
test:_FillValue = NaN ;
test:valid_range = -Infinity, Infinity ;
data:
test = -Infinity, -1.79769313486232e+308, -1, 0, 1, 2, 3, _,
1.79769313486232e+308, Infinity ;
}
✅
Summary#
The result of this experiment is underwhelming but shows a clear picture. A large set of data types can not be used reliably in a combination of netCDF and OPeNDAP. The failure modes however differ significantly between the various types:
64 bit integers simply can not be encoded in XDR which is the binary encoding of OPeNDAP. Thus the server just fails and the user receives an error.
unsigned short and unsigned int could be represented by OPeNDAP in principle, however the Server introduces spurious
_FillValues which additionally are of the wrong (signed) integral type such that the netCDF client library refuses to read the data. This is most likely a Bug in the specific server.unsigned bytes could in principle be represented by OPeNDAP, but the values received by the client are wrong. This might be an attempt by the client to somehow handle the erroneously delivered
valid_range. The big problem here is that this can result in an error without a message.signed bytes can work sometimes even if they are not representable by OPeNDAP. This is due to a hack which has been introduced into netCDF-c which is based on the use of the additional
_Unsignedattribute which is created automatically by the server. The hack however only applies to the data values and not to the attributes. As a consequence, the data type of the attributes may be changed depending on the values stored in the attributes. In particular, signed bytes seem to work only if they are positive. The behaviour is however really weird, so maybe one should not count on it.
As a consequence, the only numeric data types which should be used in any dataset are SHORT, INT, FLOAT and DOUBLE.
STRING and CHAR (when used as text) seem to be ok, but they have not been investigated in this setting yet.
Did we learn something?#
Well, it depends. In principle, these results are also present, but well hidden inside the NetCDF Users Guide: The Best Practices state:
Do not use char type variables for numeric data, use byte type variables instead.
[…]
NetCDF-3 does not have unsigned integer primitive types.
To be completely safe with unknown readers, widen the data type, or use floating point.
This excludes the use of CHAR for numeric applications and effectively also BYTE, UBYTE.
And in the data model section we can find:
For maximum interoparability with existing code, new data should be created with the The Classic Model.
This excludes the use of 64 bit integers and unsigned types.
Thus, the take-home message of this article is not new, everything could have been found in the NetCDF Users Guide. However at least I was not fully aware of all of these implications and did only find the references into the Users Guide while preparing this article. I think if we want to provide datasets which are usable by everyone, the insights of this article are important to share.
